|
Setup |
Download files required for the lesson |
| 00:00 |
1. Introduction
|
What makes research data analyses reproducible?
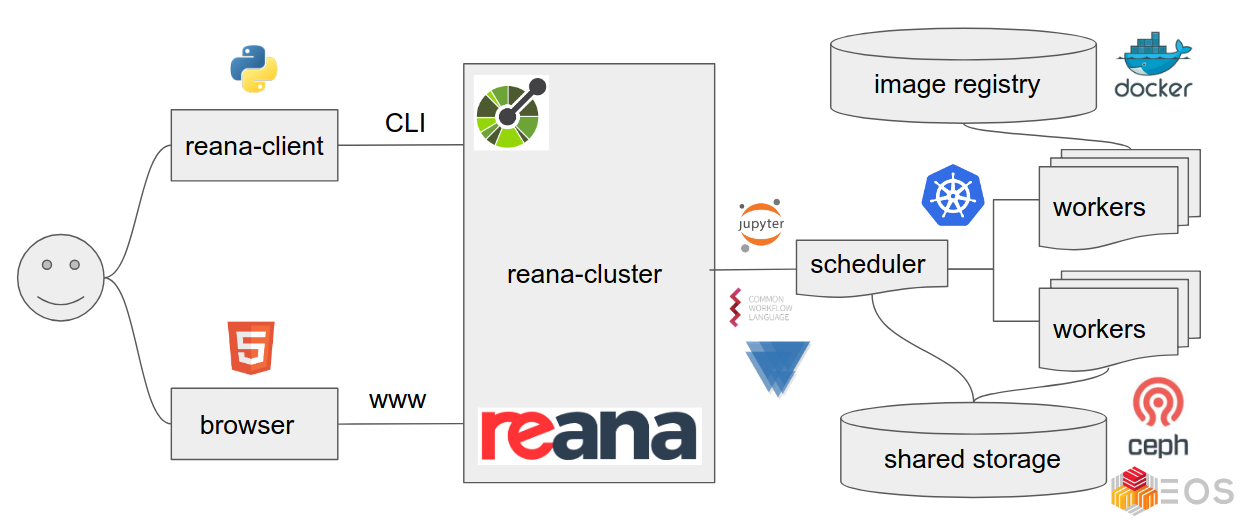
Is preserving code, data, and containers enough?
|
| 00:11 |
2. First example
|
How to run analyses on REANA cloud?
What are the basic REANA command-line client usage scenarios?
How to monitor my analysis using REANA web interface?
|
| 00:31 |
3. Developing serial workflows
|
How to write serial workflows?
What is declarative programming?
How to develop workflows progressively?
Can I temporarily override workflow parameters?
Do I always have to build new Docker image when my code changes?
|
| 01:01 |
4. HiggsToTauTau analysis: serial
|
Challenge: write the HiggsToTauTau analysis workflow and run it on REANA
|
| 01:26 |
5. Coffee break
|
Coffee break
|
| 01:41 |
6. Developing parallel workflows
|
How to scale up and run thousands of jobs?
What is a DAG?
What is a Scatter-Gather paradigm?
How to run Yadage workflows on REANA?
|
| 02:06 |
7. HiggsToTauTau analysis: parallel
|
Challenge: write the HiggsToTauTau analysis parallel workflow and run it on REANA
|
| 02:36 |
8. A glimpse on advanced topics
|
Can I publish workflow results on EOS?
Can I use Kerberos to access restricted resources?
Can I use CVMFS software repositories?
Can I dispatch heavy computations to HTCondor?
Can I dispatch heavy computations to Slurm?
Can I open Jupyter notebooks on my REANA workspace?
Can I connect my GitLab repositories with REANA?
|
| 02:56 |
9. Wrap-up
|
What have we learned today?
Where to go from here?
|
| 03:01 |
Finish |
|
The actual schedule may vary slightly depending on the topics and exercises chosen by the instructor.
 This training module is part of the HSF Software Training Center, a series of training modules that serves HEP newcomers the software skills needed as they enter the field, and in parallel, instill best practices for writing software.
This training module is part of the HSF Software Training Center, a series of training modules that serves HEP newcomers the software skills needed as they enter the field, and in parallel, instill best practices for writing software.