Getting into the Spy Game (Optional)
Overview
Teaching: 5 min
Exercises: 10 minQuestions
How can I give my GitLab CI job private information?
Objectives
Add custom environment variables
Learn how to give your CI/CD Runners access to private information
Note that you need to follow the steps in this chapter only if you are trying to use the file in CERN restricted space. If you used the file in public space you can skip to the next chapter.
So we’re nearly done with getting the merge request for the CI/CD up and running but we need to deal with this error:
$ ./skim root://eosuser.cern.ch//eos/user/g/gstark/AwesomeWorkshopFeb2020/GluGluToHToTauTau.root skim_ggH.root 19.6 11467.0 0.1
>>> Process input: root://eosuser.cern.ch//eos/user/g/gstark/AwesomeWorkshopFeb2020/GluGluToHToTauTau.root
Error in <TNetXNGFile::Open>: [ERROR] Server responded with an error: [3010] Unable to give access - user access restricted - unauthorized identity used ; Permission denied
Warning in <TTreeReader::SetEntryBase()>: There was an issue opening the last file associated to the TChain being processed.
Number of events: 0
Cross-section: 19.6
Integrated luminosity: 11467
Global scaling: 0.1
Error in <TNetXNGFile::Open>: [ERROR] Server responded with an error: [3010] Unable to give access - user access restricted - unauthorized identity used ; Permission denied
terminate called after throwing an instance of 'std::runtime_error'
what(): GetBranchNames: error in opening the tree Events
/bin/bash: line 87: 13 Aborted (core dumped) ./skim root://eosuser.cern.ch//eos/user/g/gstark/AwesomeWorkshopFeb2020/GluGluToHToTauTau.root skim_ggH.root 19.6 11467.0 0.1
section_end:1581450227:build_script
ERROR: Job failed: exit code 1
Access Control
So we need to give our CI/CD access to our data. This is actually a good thing. It means CMS can’t just grab it! Anyhow, this is pretty much done by executing printf $SERVICE_PASS | base64 -d | kinit $CERN_USER assuming that we’ve set the corresponding environment variables by safely encoding them (printf "hunter42" | base64).
Running examples with variables
Sometimes you’ll run into a code example here that you might want to run locally but relies on variables you might not have set? Sure, simply do the following
SERVICE_PASS=hunter42 CERN_USER=GoodWill printf $SERVICE_PASS | base64 -d | kinit $CERN_USER
Base-64 encoding?
Sometimes you have a string that contains certain characters that would be interpreted incorrectly by GitLab’s CI system. In order to protect against that, you can safely base-64 encode the string, store it, and then decode it as part of the CI job. This is entirely safe and recommended.
Service Account or Not?
When you’re dealing with a personal repository (project) that nobody else has administrative access to, e.g. the settings, then it’s ok to use your CERN account/password in the environment variables for the settings…
However, when you’re sharing or part of a group, it is much better to use a group’s service account or a user’s (maybe yours) service account for authentication instead. For today’s lesson however, we’ll be using your account and show pictures of how to set these environment variables.
How to make a service account?
Go to CERN Account Management -> Create New Account and click on the
Servicebutton, then clickNextand follow the steps.
Variables
There are two kinds of environment variables:
- predefined
- custom
Additionally, you can specify that the variable is a file type which is useful for passing in private keys to the CI/CD Runners. Variables can be added globally or per-job using the variables parameter.
Predefined Variables
There are quite a lot of predefined variables. We won’t cover these in depth but link for reference as they’re well-documented in the GitLab docs.
Custom Variables
Let’s go ahead and add some custom variables to fix up our access control.
- Navigate to the
Settings -> CI/CDof your repository
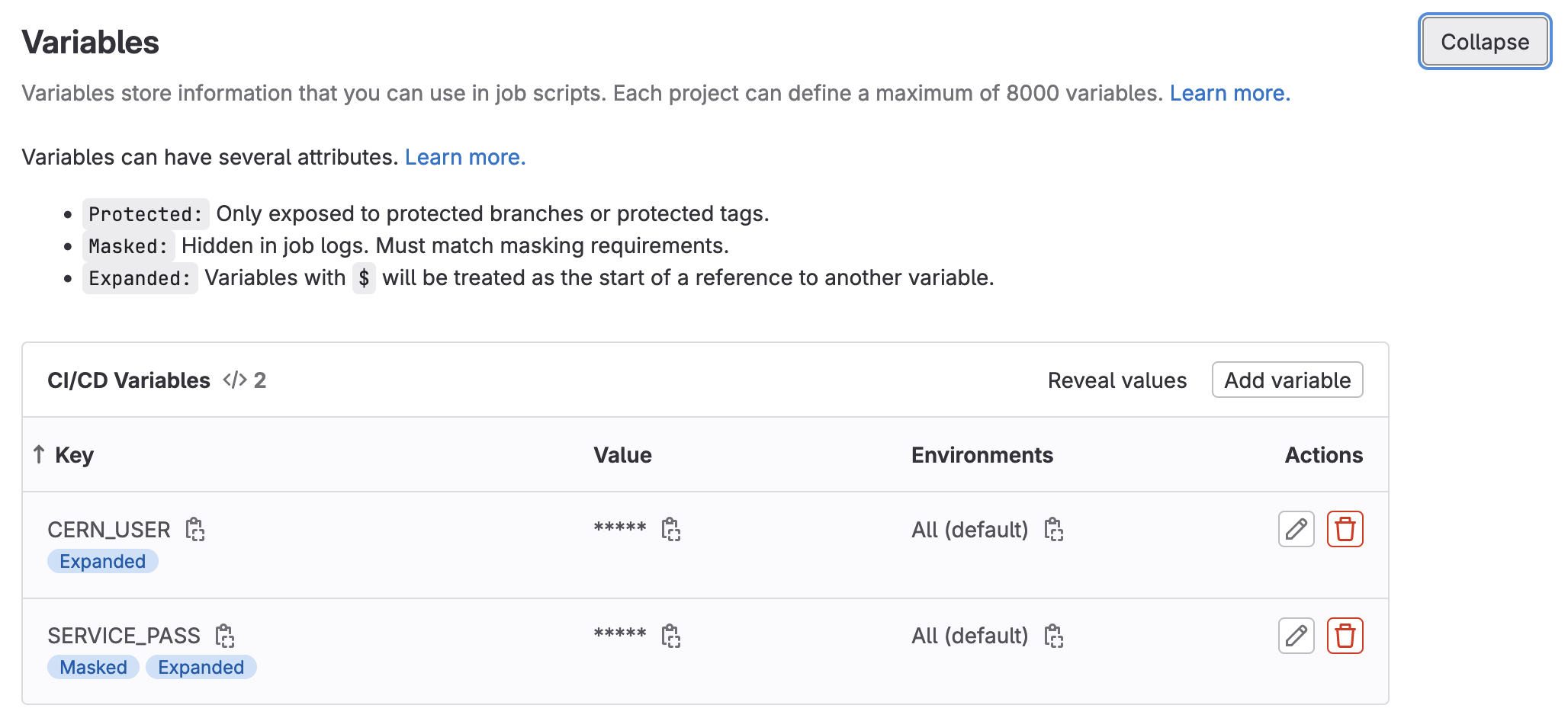
- Expand the
Variablessection on this page by clickingExpand
- Specify two environment variables,
SERVICE_PASSandCERN_USER, and fill them in appropriately. (If possible, mask the password).
Attention
- This means that we should add our
CERN_USERand ourSERVICE_PASSto the Gitlab Variables under the CI/CD section of the setting tab.- If your password is
hunter42then doprintf "hunter42" | base64 aHVudGVyNDI= # copy this as your SERVICE_PASS
- Click to save the variables.
DON’T PEEK
DON’T PEEK AT YOUR FRIEND’S SCREEN WHILE DOING THIS.
Adding kinit for access control
Now it’s time to update your CI/CD to use the environment variables you defined by adding printf $SERVICE_PASS | base64 -d | kinit $CERN_USER@CERN.CH as part of the before_script to the skim_ggH job as that’s the job that requires access.
At this point it’s also important to note that we will need a root container which has kerberos tools installed. So just for this exercise we will switch to another docker image, root:6.26.10-conda, which has those tools. In the rest of the chapters we use examples with files in public space, so you won’t need kerberos tools.
Adding Artifacts on Success
As it seems like we have a complete CI/CD that does physics - we should see what came out. We just need to add artifacts for the skim_ggH job. This is left as an exercise to you.
Adding Artifacts
Let’s add
artifactsto ourskim_ggHjob to save theskim_ggH.rootfile. Let’s have the artifacts expire in a week instead.Solution
stages: - greeting - build - run hello world: stage: greeting script: - echo "Hello World" .template_build: stage: build before_script: - COMPILER=$(root-config --cxx) - FLAGS=$(root-config --cflags --libs) script: - $COMPILER -g -O3 -Wall -Wextra -Wpedantic -o skim skim.cxx $FLAGS artifacts: paths: - skim expire_in: 1 day multi_build: extends: .template_build image: $ROOT_IMAGE parallel: matrix: - ROOT_IMAGE: ["rootproject/root:6.26.10-conda","rootproject/root:latest"] skim_ggH: stage: run dependencies: - "multi_build: [rootproject/root:6.26.10-conda]" image: rootproject/root:6.26.10-conda before_script: - printf $SERVICE_PASS | base64 -d | kinit $CERN_USER@CERN.CH script: - ./skim root://eosuser.cern.ch//eos/user/g/gstark/AwesomeWorkshopFeb2020/GluGluToHToTauTau.root skim_ggH.root 19.6 11467.0 0.1 artifacts: paths: - skim_ggH.root expire_in: 1 week
And this allows us to download artifacts from the successfully run job.
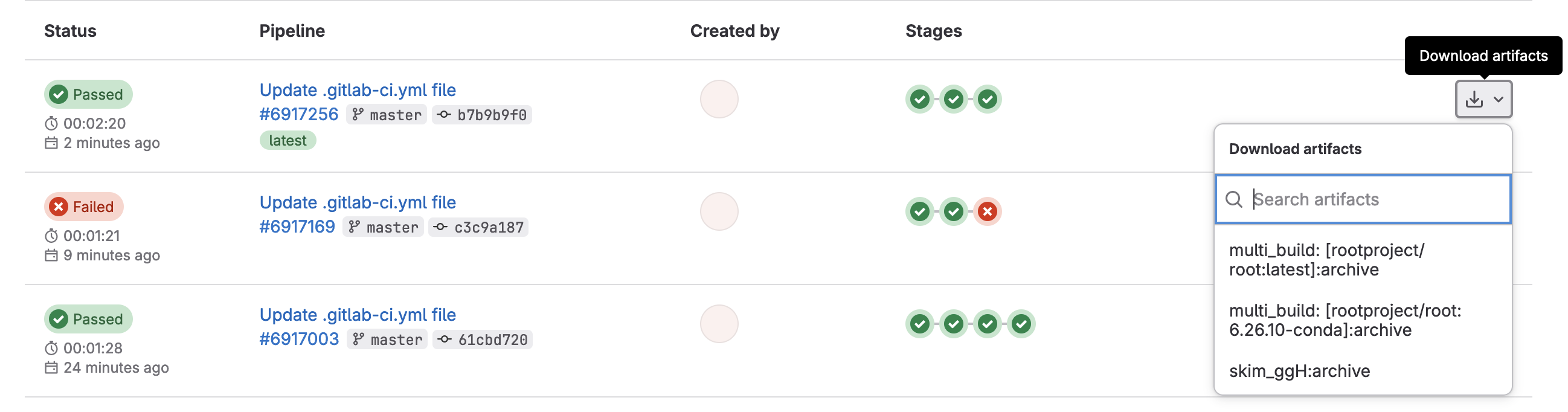
or if you click through to a skim_ggH job, you can browse the artifacts
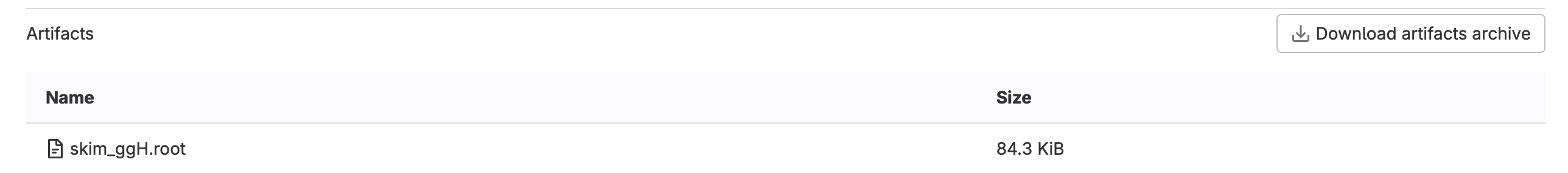
which should just be the skim_ggH.root file you just made.
Further Reading
Key Points
Service accounts provide an extra layer of security between the outside world and your account
Environment variables in GitLab CI/CD allow you to hide protected information from others who can see your code