Lesson 3: Ragged and nested arrays#
So far, all the arrays we’ve dealt with have been rectangular (in \(n\) dimensions; “rectilinear”).
What if we had data like this?
[
[[1.84, 0.324]],
[[-1.609, -0.713, 0.005], [0.953, -0.993, 0.011, 0.718]],
[[0.459, -1.517, 1.545], [0.33, 0.292]],
[[-0.376, -1.46, -0.206], [0.65, 1.278]],
[[], [], [1.617]],
[]
]
[
[[-0.106, 0.611]],
[[0.118, -1.788, 0.794, 0.658], [-0.105]]
]
[
[[-0.384], [0.697, -0.856]],
[[0.778, 0.023, -1.455, -2.289], [-0.67], [1.153, -1.669, 0.305, 1.517, -0.292]]
]
[
[[0.205, -0.355], [-0.265], [1.042]],
[[-0.004], [-1.167, -0.054, 0.726, 0.213]],
[[1.741, -0.199, 0.827]]
]
Or like this?
[
{"fill": "#b1b1b1", "stroke": "none", "points": [{"x": 5.27453, "y": 1.03276},
{"x": -3.51280, "y": 1.74849}]},
{"fill": "#b1b1b1", "stroke": "none", "points": [{"x": 8.21630, "y": 4.07844},
{"x": -0.79157, "y": 3.49478}, {"x": 16.38932, "y": 5.29399},
{"x": 10.38641, "y": 0.10832}, {"x": -2.07070, "y": 14.07140},
{"x": 9.57021, "y": -0.94823}, {"x": 1.97332, "y": 3.62380},
{"x": 5.66760, "y": 11.38001}, {"x": 0.25497, "y": 3.39276},
{"x": 3.86585, "y": 6.22051}, {"x": -0.67393, "y": 2.20572}]},
{"fill": "#d0d0ff", "stroke": "none", "points": [{"x": 3.59528, "y": 7.37191},
{"x": 0.59192, "y": 2.91503}, {"x": 4.02932, "y": -1.13601},
{"x": -1.01593, "y": 1.95894}, {"x": 1.03666, "y": 0.05251}]},
{"fill": "#d0d0ff", "stroke": "none", "points": [{"x": -8.78510, "y": -0.00497},
{"x": -15.22688, "y": 3.90244}, {"x": 5.74593, "y": 4.12718}]},
{"fill": "none", "stroke": "#000000", "points": [{"x": 4.40625, "y": -6.953125},
{"x": 4.34375, "y": -7.09375}, {"x": 4.3125, "y": -7.140625},
{"x": 4.140625, "y": -7.140625}]},
{"fill": "none", "stroke": "#808080", "points": [{"x": 0.46875, "y": -0.09375},
{"x": 0.46875, "y": -0.078125}, {"x": 0.46875, "y": 0.53125}]}
]
Or like this?
[
{"movie": "Evil Dead", "year": 1981, "actors":
["Bruce Campbell", "Ellen Sandweiss", "Richard DeManincor", "Betsy Baker"]
},
{"movie": "Darkman", "year": 1900, "actors":
["Liam Neeson", "Frances McDormand", "Larry Drake", "Bruce Campbell"]
},
{"movie": "Army of Darkness", "year": 1992, "actors":
["Bruce Campbell", "Embeth Davidtz", "Marcus Gilbert", "Bridget Fonda",
"Ted Raimi", "Patricia Tallman"]
},
{"movie": "A Simple Plan", "year": 1998, "actors":
["Bill Paxton", "Billy Bob Thornton", "Bridget Fonda", "Brent Briscoe"]
},
{"movie": "Spider-Man 2", "year": 2004, "actors":
["Tobey Maguire", "Kristen Dunst", "Alfred Molina", "James Franco",
"Rosemary Harris", "J.K. Simmons", "Stan Lee", "Bruce Campbell"]
},
{"movie": "Drag Me to Hell", "year": 2009, "actors":
["Alison Lohman", "Justin Long", "Lorna Raver", "Dileep Rao", "David Paymer"]
}
]
Or like this? (Hint: we do!)
[
{"run": 1, "luminosityBlock": 156, "event": 46501,
"PV": {"x": 0.243, "y": 0.393, "z": 1.451},
"electron": [],
"muon": [
{"pt": 63.043, "eta": -0.718, "phi": 2.968, "mass": 0.105, "charge": 1},
{"pt": 38.120, "eta": -0.879, "phi": -1.032, "mass": 0.105, "charge": -1},
{"pt": 4.048, "eta": -0.320, "phi": 1.038, "mass": 0.105, "charge": 1}
],
"MET": {"pt": 21.929, "phi": -2.730}
},
{"run": 1, "luminosityBlock": 156, "event": 46502,
"PV": {"x": 0.244, "y": 0.395, "z": -2.879},
"electron": [
{"pt": 21.902, "eta": -0.702, "phi": 0.133, "mass": 0.005, "charge": 1},
{"pt": 42.632, "eta": -0.979, "phi": -1.863, "mass": 0.008, "charge": 1},
{"pt": 78.012, "eta": -0.933, "phi": -2.207, "mass": 0.018, "charge": -1},
{"pt": 23.835, "eta": -1.362, "phi": -0.621, "mass": 0.008, "charge": -1}
],
"muon": [],
"MET": {"pt": 16.972, "phi": 2.866}}
]
It might be possible to turn these datasets into tabular form using surrogate keys and database normalization, but
they could be inconvenient or less efficient in that form, depending on what we want to do,
they were very likely given in a ragged/untidy form. You can’t ignore the data-cleaning step!
Dealing with these datasets as JSON or Python objects is inefficient for the same reason as for lists of numbers.
We want arbitrary data structure with array-oriented interface and performance…
Libraries for irregular arrays#
This is still a rather new field, but there are a few array libraries for arbitrary data structures.
Apache Arrow: In-memory format and an ecosystem of tools, an “exploded database” (database functionality provided as interchangeable pieces). Strong focus on delivering data, zero-copy, between processes.
Awkward Array: Library for array-oriented programming like NumPy, but for arbitrary data structures. Losslessly zero-copy convertible to/from Arrow and Parquet.
Parquet: Disk format for storing large datasets and (selectively) retrieving them.
ROOT RNTuple: A more flexible disk format than Parquet, in the ROOT framework.
Apache Arrow#
Arrow arrays support arbitrary data structures, but they’re intended to be used within some other interface. Increasingly, dataframe libraries are adopting Arrow as a column format.
import pyarrow as pa
arrow_array = pa.array([
[{"x": 1.1, "y": [1]}, {"x": 2.2, "y": [1, 2]}, {"x": 3.3, "y": [1, 2, 3]}],
[],
[{"x": 4.4, "y": [1, 2, 3, 4]}, {"x": 5.5, "y": [1, 2, 3, 4, 5]}]
])
arrow_array.type
ListType(list<item: struct<x: double, y: list<item: int64>>>)
arrow_array
<pyarrow.lib.ListArray object at 0x7fc50406a500>
[
-- is_valid: all not null
-- child 0 type: double
[
1.1,
2.2,
3.3
]
-- child 1 type: list<item: int64>
[
[
1
],
[
1,
2
],
[
1,
2,
3
]
],
-- is_valid: all not null
-- child 0 type: double
[]
-- child 1 type: list<item: int64>
[],
-- is_valid: all not null
-- child 0 type: double
[
4.4,
5.5
]
-- child 1 type: list<item: int64>
[
[
1,
2,
3,
4
],
[
1,
2,
3,
4,
5
]
]
]
Awkward Array#
Awkward Arrays are fully interchangeable with Arrow arrays, but they’re a high-level user interface, intended for data analysts.
import awkward as ak
awkward_array = ak.from_arrow(arrow_array)
awkward_array
[[{x: 1.1, y: [1]}, {x: 2.2, y: [...]}, {x: 3.3, y: [1, 2, 3]}],
[],
[{x: 4.4, y: [1, 2, 3, 4]}, {x: 5.5, y: [1, ..., 5]}]]
-----------------------------------------------------------------
backend: cpu
nbytes: 200 B
type: 3 * var * ?{
x: ?float64,
y: option[var * ?int64]
}Parquet#
Awkward Arrays don’t lose any information when saved as Parquet files.
ak.to_parquet(awkward_array, "/tmp/file.parquet")
<pyarrow._parquet.FileMetaData object at 0x7fc503747fb0>
created_by: parquet-cpp-arrow version 19.0.0
num_columns: 2
num_rows: 3
num_row_groups: 1
format_version: 2.6
serialized_size: 0
ak.from_parquet("/tmp/file.parquet")
[[{x: 1.1, y: [1]}, {x: 2.2, y: [...]}, {x: 3.3, y: [1, 2, 3]}],
[],
[{x: 4.4, y: [1, 2, 3, 4]}, {x: 5.5, y: [1, ..., 5]}]]
-----------------------------------------------------------------
backend: cpu
nbytes: 200 B
type: 3 * var * ?{
x: ?float64,
y: option[var * ?int64]
}RNTuple#
At the time of writing, RNTuple is very new and I can’t provide a similar example.
TO DO: Write this section when the tools are ready!
Array-oriented programming with Awkward Arrays#
Let’s start with an example to show slicing.
You can find more details in the Awkward Array documentation.
ragged = ak.Array([
[
[[1.84, 0.324]],
[[-1.609, -0.713, 0.005], [0.953, -0.993, 0.011, 0.718]],
[[0.459, -1.517, 1.545], [0.33, 0.292]],
[[-0.376, -1.46, -0.206], [0.65, 1.278]],
[[], [], [1.617]],
[]
],
[
[[-0.106, 0.611]],
[[0.118, -1.788, 0.794, 0.658], [-0.105]]
],
[
[[-0.384], [0.697, -0.856]],
[[0.778, 0.023, -1.455, -2.289], [-0.67], [1.153, -1.669, 0.305, 1.517, -0.292]]
],
[
[[0.205, -0.355], [-0.265], [1.042]],
[[-0.004], [-1.167, -0.054, 0.726, 0.213]],
[[1.741, -0.199, 0.827]]
]
])
Multidimensional indexing#
ragged[3, 1, -1, 2]
np.float64(0.726)
Range-slicing#
ragged[3, 1:, -1, 1:3]
[[-0.054, 0.726], [-0.199, 0.827]] ----------------------- backend: cpu nbytes: 56 B type: 2 * var * float64
Advanced slicing#
ragged[[False, False, True, True], [0, -1, 0, -1], 0, -1]
[-0.384, 0.827] ----------------- backend: cpu nbytes: 16 B type: 2 * float64
Awkward slicing#
Just as NumPy arrays can be sliced by NumPy arrays, Awkward Arrays can be sliced by Awkward Arrays. This lets you perform physics cuts on ragged data.
See the full Awkward slicing documentation for more.
ragged > 0
[[[[True, True]], [[False, False, True], [True, ..., True]], ..., [...], []], [[[False, True]], [[True, False, True, True], [False]]], [[[False], [True, False]], [[True, True, False, False], ..., [...]]], [[[True, False], [False], [True]], [[False], ...], [[True, False, True]]]] ----------------------------------------------------------------------------- backend: cpu nbytes: 404 B type: 4 * var * var * var * bool
ragged[ragged > 0]
[[[[1.84, 0.324]], [[0.005], [0.953, 0.011, 0.718]], [...], ..., [[], ...], []], [[[0.611]], [[0.118, 0.794, 0.658], []]], [[[], [0.697]], [[0.778, 0.023], [], [1.15, 0.305, 1.52]]], [[[0.205], [], [1.04]], [[], [0.726, 0.213]], [[1.74, 0.827]]]] -------------------------------------------------------------------------------- backend: cpu nbytes: 584 B type: 4 * var * var * var * float64
Reductions#
ak.sum(ragged)
np.float64(2.8980000000000006)
ak.sum(ragged, axis=-1)
[[[2.16], [-2.32, 0.689], [0.487, 0.622], [-2.04, ...], [0, 0, 1.62], []], [[0.505], [-0.218, -0.105]], [[-0.384, -0.159], [-2.94, -0.67, 1.01]], [[-0.15, -0.265, 1.04], [-0.004, -0.282], [2.37]]] -------------------------------------------------------------------------- backend: cpu nbytes: 344 B type: 4 * var * var * float64
ak.sum(ragged, axis=0)
[[[1.56, 0.58], [0.432, -0.856], [1.04]], [[-0.717, -2.48, -0.656, -1.63], ..., [1.15, -1.67, 0.305, 1.52, -0.292]], [[2.2, -1.72, 2.37], [0.33, 0.292]], [[-0.376, -1.46, -0.206], [0.65, 1.28]], [[], [], [1.62]], []] --------------------------------------------------------------------------- backend: cpu nbytes: 904 B type: 6 * var * var * float64
Just as regular arrays can be sliced along different axes,
ragged arrays (even with missing values like None) can be sliced by defining them to be left-aligned:
array = ak.Array([[ 1, 2, 3, 4],
[ 10, None, 30 ],
[ 100, 200 ]])
ak.sum(array, axis=0).tolist()
[111, 202, 33, 4]
ak.sum(array, axis=1).tolist()
[10, 40, 300]
In most particle physics applications, you’ll want to reduce over the deepest/maximum axis, which you can get with axis=-1.
Awkward Arrays in particle physics#
Here is a bigger version of the JSON dataset, loaded from a ROOT file with Uproot.
import uproot
file = uproot.open("data/SMHiggsToZZTo4L.root")
file
<ReadOnlyDirectory '/' at 0x7fc503f9ff90>
file.classnames()
{'Events;1': 'TTree'}
tree = file["Events"]
tree
<TTree 'Events' (32 branches) at 0x7fc52c1723d0>
tree.arrays()
[{run: 1, luminosityBlock: 156, event: 46501, PV_npvs: 15, PV_x: 0.244, ...},
{run: 1, luminosityBlock: 156, event: 46502, PV_npvs: 13, PV_x: 0.244, ...},
{run: 1, luminosityBlock: 156, event: 46503, PV_npvs: 11, PV_x: 0.242, ...},
{run: 1, luminosityBlock: 156, event: 46504, PV_npvs: 22, PV_x: 0.243, ...},
{run: 1, luminosityBlock: 156, event: 46505, PV_npvs: 18, PV_x: 0.24, ...},
{run: 1, luminosityBlock: 156, event: 46506, PV_npvs: 5, PV_x: 0.243, ...},
{run: 1, luminosityBlock: 156, event: 46507, PV_npvs: 11, PV_x: 0.242, ...},
{run: 1, luminosityBlock: 156, event: 46508, PV_npvs: 25, PV_x: 0.245, ...},
{run: 1, luminosityBlock: 156, event: 46509, PV_npvs: 12, PV_x: 0.246, ...},
{run: 1, luminosityBlock: 156, event: 46510, PV_npvs: 18, PV_x: 0.244, ...},
...,
{run: 1, luminosityBlock: 996, event: 298792, PV_npvs: 24, PV_x: 0.244, ...},
{run: 1, luminosityBlock: 996, event: 298793, PV_npvs: 14, PV_x: 0.242, ...},
{run: 1, luminosityBlock: 996, event: 298794, PV_npvs: 24, PV_x: 0.245, ...},
{run: 1, luminosityBlock: 996, event: 298795, PV_npvs: 30, PV_x: 0.245, ...},
{run: 1, luminosityBlock: 996, event: 298796, PV_npvs: 15, PV_x: 0.244, ...},
{run: 1, luminosityBlock: 996, event: 298797, PV_npvs: 18, PV_x: 0.243, ...},
{run: 1, luminosityBlock: 996, event: 298798, PV_npvs: 9, PV_x: 0.245, ...},
{run: 1, luminosityBlock: 996, event: 298799, PV_npvs: 15, PV_x: 0.243, ...},
{run: 1, luminosityBlock: 996, event: 298800, PV_npvs: 13, PV_x: 0.247, ...}]
-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
backend: cpu
nbytes: 101.2 MB
type: 299973 * {
run: int32,
luminosityBlock: uint32,
event: uint64,
PV_npvs: int32,
PV_x: float32,
PV_y: float32,
PV_z: float32,
nMuon: uint32,
Muon_pt: var * float32,
Muon_eta: var * float32,
Muon_phi: var * float32,
Muon_mass: var * float32,
Muon_charge: var * int32,
Muon_pfRelIso03_all: var * float32,
Muon_pfRelIso04_all: var * float32,
Muon_dxy: var * float32,
Muon_dxyErr: var * float32,
Muon_dz: var * float32,
Muon_dzErr: var * float32,
nElectron: uint32,
Electron_pt: var * float32,
Electron_eta: var * float32,
Electron_phi: var * float32,
Electron_mass: var * float32,
Electron_charge: var * int32,
Electron_pfRelIso03_all: var * float32,
Electron_dxy: var * float32,
Electron_dxyErr: var * float32,
Electron_dz: var * float32,
Electron_dzErr: var * float32,
MET_pt: float32,
MET_phi: float32
}Here it is again (with more JSON-like formatting) from a Parquet file.
events = ak.from_parquet("data/SMHiggsToZZTo4L.parquet")
events
[{run: 1, luminosityBlock: 156, event: 46501, PV: {...}, electron: [], ...},
{run: 1, luminosityBlock: 156, event: 46502, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 156, event: 46503, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 156, event: 46504, PV: {...}, electron: ..., ...},
{run: 1, luminosityBlock: 156, event: 46505, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 156, event: 46506, PV: {...}, electron: ..., ...},
{run: 1, luminosityBlock: 156, event: 46507, PV: {...}, electron: ..., ...},
{run: 1, luminosityBlock: 156, event: 46508, PV: {...}, electron: ..., ...},
{run: 1, luminosityBlock: 156, event: 46509, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 156, event: 46510, PV: {...}, electron: [], ...},
...,
{run: 1, luminosityBlock: 996, event: 298792, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 996, event: 298793, PV: {...}, electron: ..., ...},
{run: 1, luminosityBlock: 996, event: 298794, PV: {...}, electron: [], ...},
{run: 1, luminosityBlock: 996, event: 298795, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 996, event: 298796, PV: {...}, electron: [], ...},
{run: 1, luminosityBlock: 996, event: 298797, PV: {...}, electron: ..., ...},
{run: 1, luminosityBlock: 996, event: 298798, PV: {...}, electron: [], ...},
{run: 1, luminosityBlock: 996, event: 298799, PV: {...}, electron: [...], ...},
{run: 1, luminosityBlock: 996, event: 298800, PV: {...}, electron: [...], ...}]
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
backend: cpu
nbytes: 53.2 MB
type: 299973 * {
run: int32,
luminosityBlock: int64,
event: uint64,
PV: Vector3D[
x: float32,
y: float32,
z: float32
],
electron: var * Momentum4D[
pt: float32,
eta: float32,
phi: float32,
mass: float32,
charge: int32,
pfRelIso03_all: float32,
dxy: float32,
dxyErr: float32,
dz: float32,
dzErr: float32
],
muon: var * Momentum4D[
pt: float32,
eta: float32,
phi: float32,
mass: float32,
charge: int32,
pfRelIso03_all: float32,
pfRelIso04_all: float32,
dxy: float32,
dxyErr: float32,
dz: float32,
dzErr: float32
],
MET: Momentum2D[
pt: float32,
phi: float32
]
}View the first event as Python lists and dicts (like JSON).
events[0].to_list()
{'run': 1,
'luminosityBlock': 156,
'event': 46501,
'PV': {'x': 0.24369880557060242,
'y': 0.3936990201473236,
'z': 1.451307773590088},
'electron': [],
'muon': [{'pt': 63.04386901855469,
'eta': -0.7186822295188904,
'phi': 2.968005895614624,
'mass': 0.10565836727619171,
'charge': 1,
'pfRelIso03_all': 0.0,
'pfRelIso04_all': 0.0,
'dxy': -0.004785160068422556,
'dxyErr': 0.0060764215886592865,
'dz': 0.09005985409021378,
'dzErr': 0.044572051614522934},
{'pt': 38.12034606933594,
'eta': -0.8794569969177246,
'phi': -1.0324749946594238,
'mass': 0.10565836727619171,
'charge': -1,
'pfRelIso03_all': 0.0,
'pfRelIso04_all': 0.0,
'dxy': 0.0005746808601543307,
'dxyErr': 0.0013040687190368772,
'dz': -0.0032290113158524036,
'dzErr': 0.003023269586265087},
{'pt': 4.04868745803833,
'eta': -0.320764422416687,
'phi': 1.0385035276412964,
'mass': 0.10565836727619171,
'charge': 1,
'pfRelIso03_all': 0.0,
'pfRelIso04_all': 0.17997965216636658,
'dxy': -0.00232272082939744,
'dxyErr': 0.004343290813267231,
'dz': -0.005162843037396669,
'dzErr': 0.004190043080598116}],
'MET': {'pt': 21.929929733276367, 'phi': -2.7301223278045654}}
Get one numeric field (also known as “column”).
events.electron.pt
[[], [21.9, 42.6, 78, 23.8], [11.6, 6.69], [10.4], [30.7, 29.2, 6.38, 6.24], [16.1], [5.32], [6.99], [41, 6.5, 13.1, 52.1], [], ..., [19.1, 9.69], [30.2], [], [37, 7.36, 48], [], [12.3], [], [17.9, 23.2], [48.1, 38.7]] ---------------------------- backend: cpu nbytes: 4.1 MB type: 299973 * var * float32
Compute something (\(p_z = p_T \sinh\eta\)).
import numpy as np
events.electron.pt * np.sinh(events.electron.eta)
[[], [-16.7, -48.8, -83.9, -43.5], [-11.1, 26], [-0.237], [-19.9, 47.5, -18, -15.1], [5.58], [-3.22], [3.88], [-8.92, -10.5, -23.8, -17.3], [], ..., [26.4, 19.1], [92.2], [], [-193, -2.78, -43.4], [], [-3.4], [], [80.1, 99.3], [26.8, 74]] ------------------------------ backend: cpu nbytes: 4.1 MB type: 299973 * var * float32
The Vector library also has an Awkward Array backend, which is documented here. To use it, call register_awkward after importing the library.
import vector
vector.register_awkward()
Now records with name="Momentum4D" and fields with coordinate names (px, py, pz, E or pt, phi, eta, m) automatically get Vector properties and methods.
events.electron.type.show()
299973 * var * Momentum4D[
pt: float32,
eta: float32,
phi: float32,
mass: float32,
charge: int32,
pfRelIso03_all: float32,
dxy: float32,
dxyErr: float32,
dz: float32,
dzErr: float32
]
# implicitly computes pz = pt * sinh(eta)
events.electron.pz
[[], [-16.7, -48.8, -83.9, -43.5], [-11.1, 26], [-0.237], [-19.9, 47.5, -18, -15.1], [5.58], [-3.22], [3.88], [-8.92, -10.5, -23.8, -17.3], [], ..., [26.4, 19.1], [92.2], [], [-193, -2.78, -43.4], [], [-3.4], [], [80.1, 99.3], [26.8, 74]] ------------------------------ backend: cpu nbytes: 4.1 MB type: 299973 * var * float32
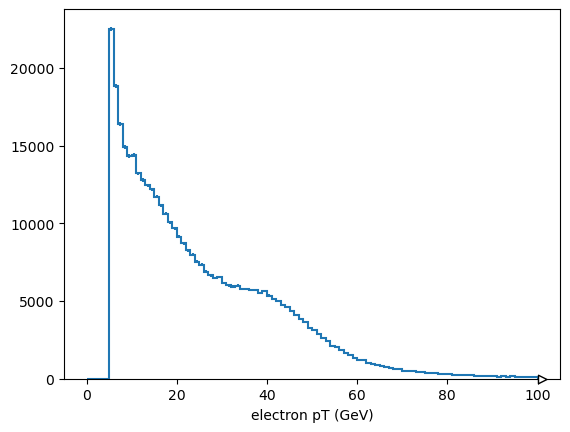
To make histograms or other plots, we need numbers without structure, so ak.flatten the array.
from hist import Hist
Hist.new.Regular(100, 0, 100, name="electron pT (GeV)").Double().fill(
ak.flatten(events.electron.pt)
).plot();
Each event has a different number of electrons and muons (ak.num to check).
ak.num(events.electron), ak.num(events.muon)
(<Array [0, 4, 2, 1, 4, 1, 1, 1, ..., 0, 3, 0, 1, 0, 2, 2] type='299973 * int64'>,
<Array [3, 0, 0, 7, 0, 2, 1, 0, ..., 2, 0, 2, 2, 4, 0, 0] type='299973 * int64'>)
So what happens if we try to compute something with the electrons’ \(p_T\) and the muons’ \(\eta\)?
events.electron.pt * np.sinh(events.muon.eta)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[37], line 1
----> 1 events.electron.pt * np.sinh(events.muon.eta)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_operators.py:54, in _binary_method.<locals>.func(self, other)
51 if _disables_array_ufunc(other):
52 return NotImplemented
---> 54 return ufunc(self, other)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/highlevel.py:1619, in Array.__array_ufunc__(self, ufunc, method, *inputs, **kwargs)
1554 """
1555 Intercepts attempts to pass this Array to a NumPy
1556 [universal functions](https://docs.scipy.org/doc/numpy/reference/ufuncs.html)
(...)
1616 See also #__array_function__.
1617 """
1618 name = f"{type(ufunc).__module__}.{ufunc.__name__}.{method!s}"
-> 1619 with ak._errors.OperationErrorContext(name, inputs, kwargs):
1620 return ak._connect.numpy.array_ufunc(ufunc, method, inputs, kwargs)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_errors.py:80, in ErrorContext.__exit__(self, exception_type, exception_value, traceback)
78 self._slate.__dict__.clear()
79 # Handle caught exception
---> 80 raise self.decorate_exception(exception_type, exception_value)
81 else:
82 # Step out of the way so that another ErrorContext can become primary.
83 if self.primary() is self:
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/highlevel.py:1620, in Array.__array_ufunc__(self, ufunc, method, *inputs, **kwargs)
1618 name = f"{type(ufunc).__module__}.{ufunc.__name__}.{method!s}"
1619 with ak._errors.OperationErrorContext(name, inputs, kwargs):
-> 1620 return ak._connect.numpy.array_ufunc(ufunc, method, inputs, kwargs)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_connect/numpy.py:469, in array_ufunc(ufunc, method, inputs, kwargs)
461 raise TypeError(
462 "no {}.{} overloads for custom types: {}".format(
463 type(ufunc).__module__, ufunc.__name__, ", ".join(error_message)
464 )
465 )
467 return None
--> 469 out = ak._broadcasting.broadcast_and_apply(
470 inputs,
471 action,
472 depth_context=depth_context,
473 lateral_context=lateral_context,
474 allow_records=False,
475 function_name=ufunc.__name__,
476 )
478 out_named_axis = functools.reduce(
479 _unify_named_axis, lateral_context[NAMED_AXIS_KEY].named_axis
480 )
481 if len(out) == 1:
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:1200, in broadcast_and_apply(inputs, action, depth_context, lateral_context, allow_records, left_broadcast, right_broadcast, numpy_to_regular, regular_to_jagged, function_name, broadcast_parameters_rule)
1198 backend = backend_of(*inputs, coerce_to_common=False)
1199 isscalar = []
-> 1200 out = apply_step(
1201 backend,
1202 broadcast_pack(inputs, isscalar),
1203 action,
1204 0,
1205 depth_context,
1206 lateral_context,
1207 {
1208 "allow_records": allow_records,
1209 "left_broadcast": left_broadcast,
1210 "right_broadcast": right_broadcast,
1211 "numpy_to_regular": numpy_to_regular,
1212 "regular_to_jagged": regular_to_jagged,
1213 "function_name": function_name,
1214 "broadcast_parameters_rule": broadcast_parameters_rule,
1215 },
1216 )
1217 assert isinstance(out, tuple)
1218 return tuple(broadcast_unpack(x, isscalar) for x in out)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:1178, in apply_step(backend, inputs, action, depth, depth_context, lateral_context, options)
1176 return result
1177 elif result is None:
-> 1178 return continuation()
1179 else:
1180 raise AssertionError(result)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:1147, in apply_step.<locals>.continuation()
1145 # Any non-string list-types?
1146 elif any(x.is_list and not is_string_like(x) for x in contents):
-> 1147 return broadcast_any_list()
1149 # Any RecordArrays?
1150 elif any(x.is_record for x in contents):
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:671, in apply_step.<locals>.broadcast_any_list()
668 nextinputs.append(x)
669 nextparameters.append(NO_PARAMETERS)
--> 671 outcontent = apply_step(
672 backend,
673 nextinputs,
674 action,
675 depth + 1,
676 copy.copy(depth_context),
677 lateral_context,
678 options,
679 )
680 assert isinstance(outcontent, tuple)
681 parameters = parameters_factory(nextparameters, len(outcontent))
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:1178, in apply_step(backend, inputs, action, depth, depth_context, lateral_context, options)
1176 return result
1177 elif result is None:
-> 1178 return continuation()
1179 else:
1180 raise AssertionError(result)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:1147, in apply_step.<locals>.continuation()
1145 # Any non-string list-types?
1146 elif any(x.is_list and not is_string_like(x) for x in contents):
-> 1147 return broadcast_any_list()
1149 # Any RecordArrays?
1150 elif any(x.is_record for x in contents):
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:722, in apply_step.<locals>.broadcast_any_list()
718 for i, ((named_axis, ndim), x, x_is_string) in enumerate(
719 zip(named_axes_with_ndims, inputs, input_is_string)
720 ):
721 if isinstance(x, listtypes) and not x_is_string:
--> 722 next_content = broadcast_to_offsets_avoiding_carry(x, offsets)
723 nextinputs.append(next_content)
724 nextparameters.append(x._parameters)
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/_broadcasting.py:385, in broadcast_to_offsets_avoiding_carry(list_content, offsets)
383 return list_content.content[:next_length]
384 else:
--> 385 return list_content._broadcast_tooffsets64(offsets).content
386 elif isinstance(list_content, ListArray):
387 # Is this list contiguous?
388 if index_nplike.array_equal(
389 list_content.starts.data[1:], list_content.stops.data[:-1]
390 ):
391 # Does this list match the offsets?
File ~/micromamba/envs/array-oriented-programming/lib/python3.11/site-packages/awkward/contents/listoffsetarray.py:423, in ListOffsetArray._broadcast_tooffsets64(self, offsets)
418 next_content = self._content[this_start:]
420 if index_nplike.known_data and not index_nplike.array_equal(
421 this_zero_offsets, offsets.data
422 ):
--> 423 raise ValueError("cannot broadcast nested list")
425 return ListOffsetArray(
426 offsets, next_content[: offsets[-1]], parameters=self._parameters
427 )
ValueError: cannot broadcast nested list
This error occurred while calling
numpy.multiply.__call__(
<Array [[], [21.9, ...], ..., [48.1, 38.7]] type='299973 * var * fl...'>
<Array [[-0.782, -0.997, -0.326], ..., []] type='299973 * var * flo...'>
)
This is data structure-aware, array-oriented programming.
Application: Filtering events with an array of booleans#
events.MET.pt > 20 is a boolean array with the same length as events, so applying it as a slice selects events with MET \(p_T > 20\) GeV.
events.MET.pt, events.MET.pt > 20
(<Array [21.9, 17, 19.1, 30.9, ..., 17.7, 24, 12.9] type='299973 * float32'>,
<Array [True, False, False, True, ..., False, True, False] type='299973 * bool'>)
After slicing, the number of events is smaller.
len(events), len(events[events.MET.pt > 20])
(299973, 163222)
Application: Filtering particles with an array of lists of booleans#
events.electron.pt > 30 is a ragged boolean array with the same length-per-list as events.electron. Applying it as a slice selects electrons with \(p_T > 30\) GeV.
events.electron.pt, events.electron.pt > 30
(<Array [[], [21.9, ..., 23.8], ..., [48.1, 38.7]] type='299973 * var * float32'>,
<Array [[], [False, ..., False], ..., [True, True]] type='299973 * var * bool'>)
After slicing, the number of electrons in each event can be smaller.
ak.num(events.electron), ak.num(events.electron[events.electron.pt > 30])
(<Array [0, 4, 2, 1, 4, 1, 1, 1, ..., 0, 3, 0, 1, 0, 2, 2] type='299973 * int64'>,
<Array [0, 2, 0, 0, 1, 0, 0, 0, ..., 0, 2, 0, 0, 0, 0, 2] type='299973 * int64'>)
Mini-quiz 1: Using the reducer ak.any, how would we select events in which any electron has \(p_T > 30\) GeV/c\(^2\)?
Ragged combinatorics#
Awkward Array has two combinatorial primitives:
ak.cartesian takes a Cartesian product of lists from \(N\) different arrays, producing an array of lists of \(N\)-tuples.
ak.combinations takes \(N\) samples without replacement of lists from a single array, producing an array of lists of \(N\)-tuples.
numbers = ak.Array([[1, 2, 3], [], [4]])
letters = ak.Array([["a", "b"], ["c"], ["d", "e"]])
ak.cartesian([numbers, letters])
[[(1, 'a'), (1, 'b'), (2, 'a'), (2, 'b'), (3, 'a'), (3, 'b')],
[],
[(4, 'd'), (4, 'e')]]
--------------------------------------------------------------
backend: cpu
nbytes: 229 B
type: 3 * var * (
int64,
string
)values = ak.Array([[1.1, 2.2, 3.3, 4.4], [], [5.5, 6.6]])
ak.combinations(values, 2)
[[(1.1, 2.2), (1.1, 3.3), (1.1, 4.4), (2.2, ...), (2.2, 4.4), (3.3, 4.4)],
[],
[(5.5, 6.6)]]
--------------------------------------------------------------------------
backend: cpu
nbytes: 144 B
type: 3 * var * (
float64,
float64
)Often, it’s useful to separate the separate the left-hand sides and right-hand sides of these pairs with ak.unzip, so they can be used in mathematical expressions.
electron_muon_pairs = ak.cartesian([events.electron, events.muon])
electron_in_pair, muon_in_pair = ak.unzip(electron_muon_pairs)
electron_in_pair.pt, muon_in_pair.pt
(<Array [[], [], [], [10.4, ...], ..., [], [], []] type='299973 * var * float32'>,
<Array [[], [], [], [54.3, ...], ..., [], [], []] type='299973 * var * float32'>)
ak.num(electron_in_pair), ak.num(muon_in_pair)
(<Array [0, 0, 0, 7, 0, 2, 1, 0, ..., 0, 0, 0, 2, 0, 0, 0] type='299973 * int64'>,
<Array [0, 0, 0, 7, 0, 2, 1, 0, ..., 0, 0, 0, 2, 0, 0, 0] type='299973 * int64'>)
To use Vector’s deltaR method (\(\Delta R = \sqrt{\Delta\phi^2 + \Delta\eta^2}\)), we need to have the electrons and muons in separate arrays.
electron_in_pair, muon_in_pair = ak.unzip(ak.cartesian([events.electron, events.muon]))
electron_in_pair.deltaR(muon_in_pair)
[[], [], [], [1.07, 0.405, 1.97, 2.78, 1.01, 0.906, 2.79], [], [2.55, 0.8], [2.63], [], [], [], ..., [0.44, 3.16, 3.01, 1.05], [3.06, 1.52], [], [], [], [2.54, 0.912], [], [], []] ---------------------------------------------- backend: cpu nbytes: 3.9 MB type: 299973 * var * float32
Same for combinations from a single collection.
first_electron_in_pair, second_electron_in_pair = ak.unzip(ak.combinations(events.electron, 2))
first_electron_in_pair.deltaR(second_electron_in_pair)
[[], [2.02, 2.35, 1, 0.347, 1.3, 1.64], [2.96], [], [3.31, 1.23, 2.39, 3.8, 3.19, 2.61], [], [], [], [1.18, 2.05, 2.98, 2.26, 2.59, 1.9], [], ..., [2.87], [], [], [2.05, 2.92, 3.02], [], [], [], [2.96], [2.72]] ------------------------------------- backend: cpu nbytes: 3.7 MB type: 299973 * var * float32
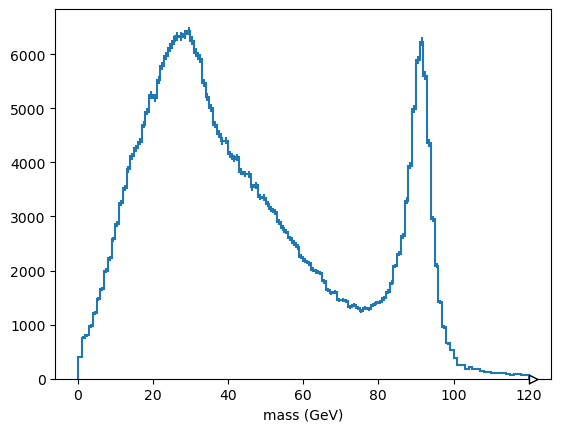
Application: Z mass computed from all combinations of electrons#
A rough Z mass plot can be made in three lines of code.
first_electron_in_pair, second_electron_in_pair = ak.unzip(ak.combinations(events.electron, 2))
z_mass = (first_electron_in_pair + second_electron_in_pair).mass
Hist.new.Reg(120, 0, 120, name="mass (GeV)").Double().fill(
ak.flatten(z_mass, axis=-1)
).plot();
Lesson 3 project: Higgs combinatorics from arrays#
As described in the intro, navigate to the notebooks directory and open lesson-3-project.ipynb, then follow its instructions.